Energetic Fingerprinting
Many enzymes of therapeutic importance, such as protein kinases that use common cofactors like ATP, were long considered to be unsuitable drug discovery targets since it was anticipated that it would not be possible to make analogs that would bind to the ATP binding site of the target enzyme with sufficient specificity to differentiate the drug action from related kinases. Such situations can be more quantitatively characterized by screening a library of structurally related analogs (e.g. ATP analogs and substructures for the kinase example illustrated) using thermofluor to differentiate the ligand binding characteristics of the different binding sites.
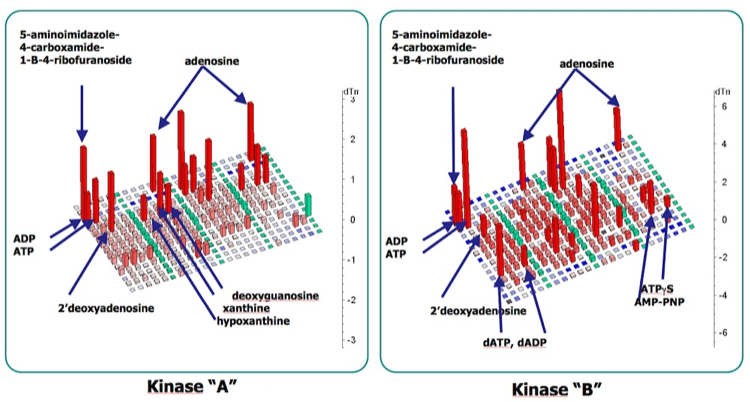
Thermofluor of ATP analogs binding to different Tyrosine Kinases Domains