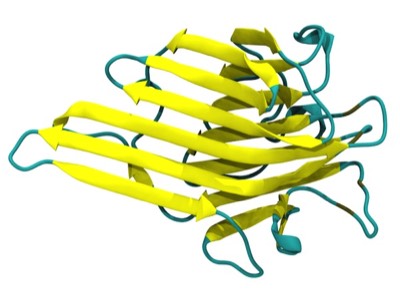
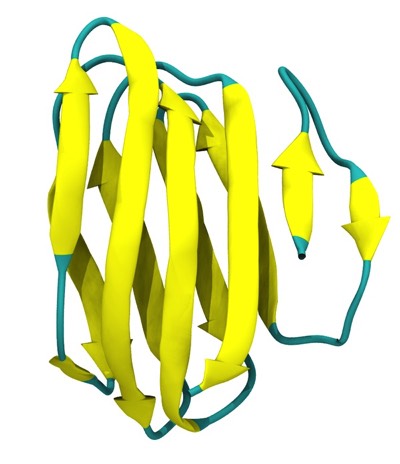
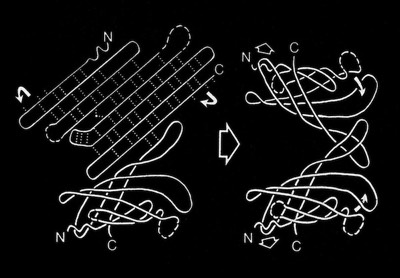
Beta Sheets
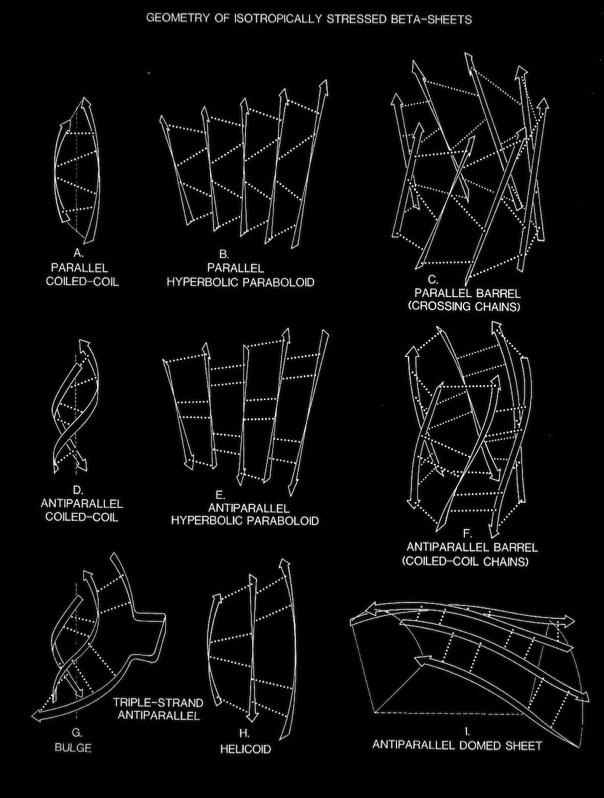
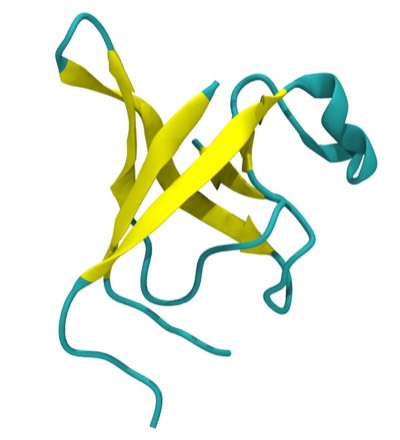
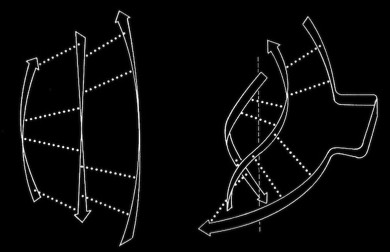
Beta sheets and alpha helices represent the major classes of extended hydrogen-bonded secondary structures found in proteins. One of the primary structural observations to emerge from early protein X-ray structures was the right-hand “twisted” character of protein beta sheets. Although the alpha-helices in globular proteins like myoglobin looked essentially like the structures predicted from basic helical polypeptide models, beta sheets in globular proteins were found to have a wide range of extended geometries, although all seemed to share a “right-handed” twist. These structures included unusually curved surfaces and cyclically H-bonded barrels that were very different from the flat beta sheet models then proposed for structural proteins like the beta-keratins and silk.
Dave Weatherford and I were able to compute coordinates for atomic models that closely approximated the observed sheet configurations. We were also able to demonstrate how basic constraints on the geometry of interchain H-bonding resulted in the many unusual (at the time) observed beta sheet geometries and why such structures might be expected to recur in proteins lacking common sequence features.
At the time we began our work (1977), there were 36 coordinate entries in the Brookhaven Protein Data Bank, and tools for protein structural modeling and representational graphics were in their infancy. We built custom codes to generate our model structures and illustrated them with India ink tracings of ORTEP plots. Subsequent work in later years by others would have the benefit of much improved analytical and graphics tools, as well as a greatly expanded database of observed structures from which to build and extend structural classifications.
At the time we began our work (1977), there were 36 coordinate entries in the Brookhaven Protein Data Bank, and tools for protein structural modeling and representational graphics were in their infancy. We built custom codes to generate our model structures and illustrated them with India ink tracings of ORTEP plots. Subsequent work in later years by others would have the benefit of much improved analytical and graphics tools, as well as a greatly expanded database of observed structures from which to build and extend structural classifications.
Real beta sheets observed in protein structures seldom show the perfect phi, psi regularity of the models that we developed, but rather appear to reflect local conformational and packing adjustments that are associated with the final, optimal packing of amino acid side chains in folded protein’s interior. Most importantly, our work showed that partially or completely assembled sheets could cooperatively deform to form final structures that corresponded to minimal surfaces, thus defining plausible structured intermediates that could form prior to the final steps in protein folding, that essentially involve aperiodic crystallization of the amino acid side chains in the protein interior.
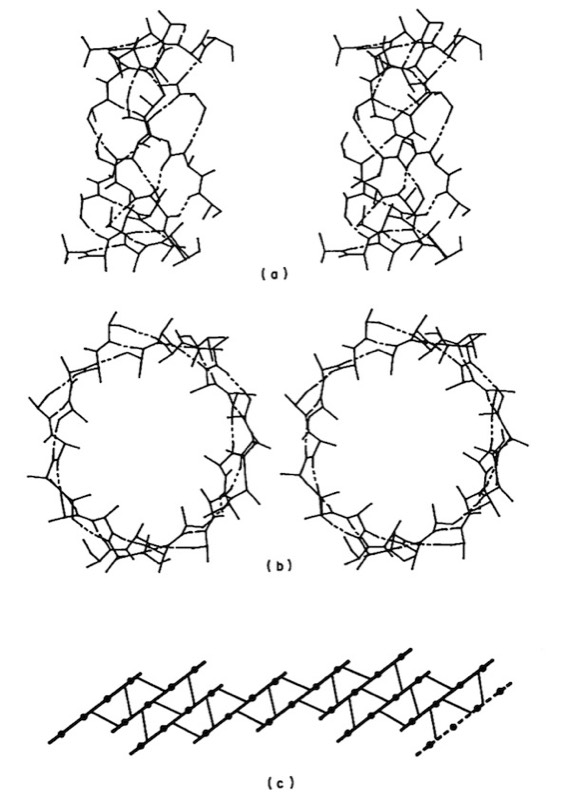
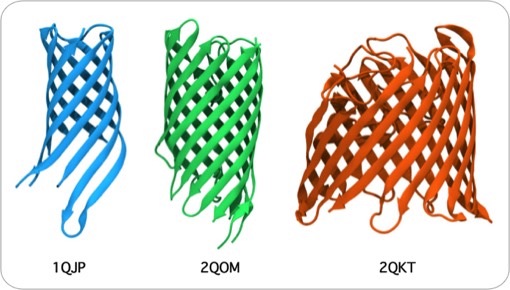
Overall, our work provided the first quantitative descriptions and physical explanations of important features of beta sheet structure in globular proteins, and anticipated many kinds of structures, such as antiparallel beta barrels with variable diameters composed of coiled polypeptide chains, that would only be structurally characterized using X-ray crystallography many years later.
Overall, our work provided the first quantitative descriptions and physical explanations of important features of beta sheet structure in globular proteins, and anticipated many kinds of structures, such as antiparallel beta barrels with variable diameters composed of coiled polypeptide chains, that would only be structurally characterized using X-ray crystallography many years later.
Antiparallel Beta Barrels Formed of Coiled Polypeptide Conformations
Beta Sheet Polypeptide Conformation
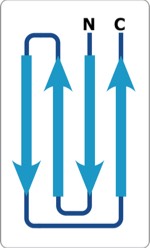
Beta sheets are composed of polypeptide chains with 2(1) screw axes of symmetry relating consecutive residues of the polypeptide chains. Parallel sheets incorporate adjacent H-bonded polypeptide chains running in the same N to C direction, creating an additional 2(1) screw axis midway between adjacent chains of the sheet. Antiparallel sheets incorporate adjacent H-bonded polypeptide chains running in opposite N to C directions, creating structures with alternating “small” and “large” H-bonded rings and additional dyad symmetry axes in the center of each ring normal to the sheet plane.
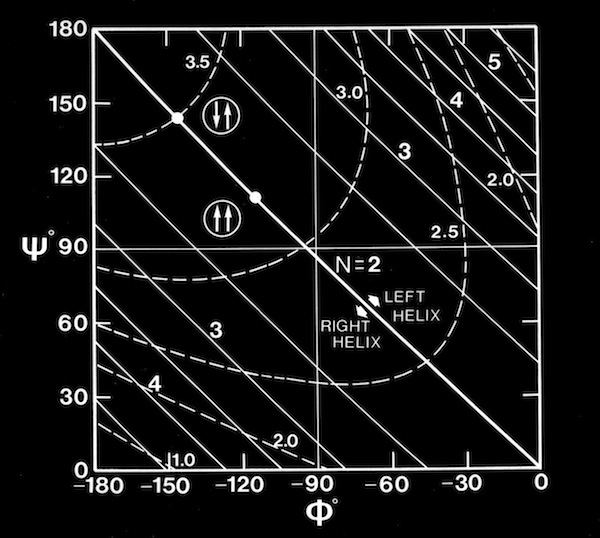
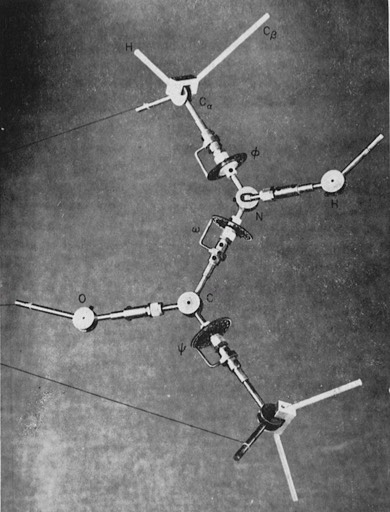
The conformational properties of polypeptides had been studied in detail by Ramachandran. He assumed planar trans-peptide bonds, and consequently was able to defined sterically allowed protein secondary structures in terms of the peptide backbone torsions (phi and psi) around the C(alpha)-C and C(alpha)-N bonds of the polypeptide backbone. It quickly became customary to plot phi,psi angles calculated from newly determined protein structures on a “Ramachandran plot” to check for conformational anomalies, although some unexpected ones (e.g. cis-peptide bonds) do exist in protein structures. Nevertheless, it was clear from inspection of Ramachandran plots of twisted beta sheets that the polypeptide chains generally had phi,psi angles corresponding to chains with a local left-handed helical twist, which for essentially extended polypeptide chains, produces sheets with a right-handed twist.
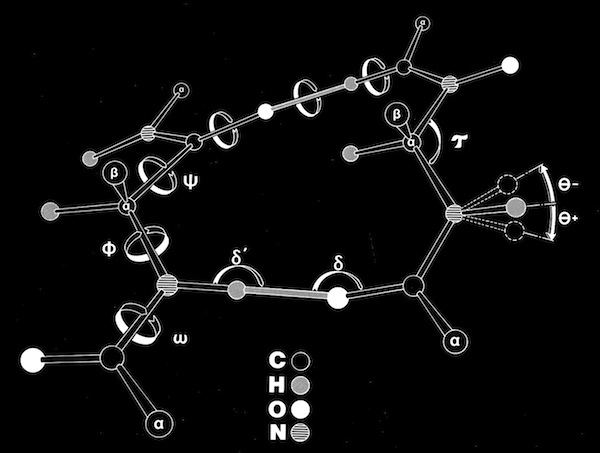
As an assistant professor of Chemistry at the University of Arizona, I constructed some very large scale (10 cm/Angstroms) peptide models that incorporated both the canonical phi-psi peptide degrees of freedom, as well as additional degrees of freedom, including peptide bond nonplanarity and the ability of the peptide amide nitrogen to assume some tetrahedral sp3 character, as predicted from some theoretical studies.
Polypeptide Model Adjustable Geometry
I used the large-scale models to build some large parallel and antiparallel beta sheets (about 2-3 meters on a side!). The models incorporated fixed-length H-O H-bonds, but allowed H-bonds to be formed along any vector lying in a plane defined by the sp2 orbital of the peptide carbonyl oxygen, as consistent with then current theory. Dial indicators on all of the phi and psi angles gave a continuous readout when the structures were manipulated. A number of interesting observations emerged.
Large parallel sheets, constructed with straight H-Bonds, could be smoothly twisted (in both left and right handed directions) in a completely cooperative and reproducible manner. At the time, this struck me as a mechanical marvel for such an extensively coupled structure of non-linear crankshafts (a poorly studied class of mechanical linkages). Equally important, the ability to flexibly twist was abruptly limited at the most extreme extent of twist, which was basically determined by the overall extent and plan of H-bonds across the sheet.
Mechanical models of antiparallel sheets were generally much more flexible, both because the dipeptide repeat period was variable, and because the antiparallel sheets could accommodate highly coiled polypeptide chain conformations.
At that point (circa 1976) I was joined by David Weatherford, an extremely talented programmer who was entering the UAZ chemistry PhD program. David was basically able to embody all of the features of the mechanical models into a computational system for building beta sheets, whose properties were ultimately constrained by the requirement to maintain low energy inter-chain hydrogen-bonds. This subsequently allowed us to produce atomic models of twisted sheets, typically incorporating polypeptide chains with completely regular phi,psi conformational angles. In a great many cases, these models corresponded very closely (C(alpha) RMS fits < 1 Angstrom) to the coordinates of structures determined from X-Ray crystallography.
Mechanical models of antiparallel sheets were generally much more flexible, both because the dipeptide repeat period was variable, and because the antiparallel sheets could accommodate highly coiled polypeptide chain conformations.
At that point (circa 1976) I was joined by David Weatherford, an extremely talented programmer who was entering the UAZ chemistry PhD program. David was basically able to embody all of the features of the mechanical models into a computational system for building beta sheets, whose properties were ultimately constrained by the requirement to maintain low energy inter-chain hydrogen-bonds. This subsequently allowed us to produce atomic models of twisted sheets, typically incorporating polypeptide chains with completely regular phi,psi conformational angles. In a great many cases, these models corresponded very closely (C(alpha) RMS fits < 1 Angstrom) to the coordinates of structures determined from X-Ray crystallography.
Parallel Beta-Sheet Twisting
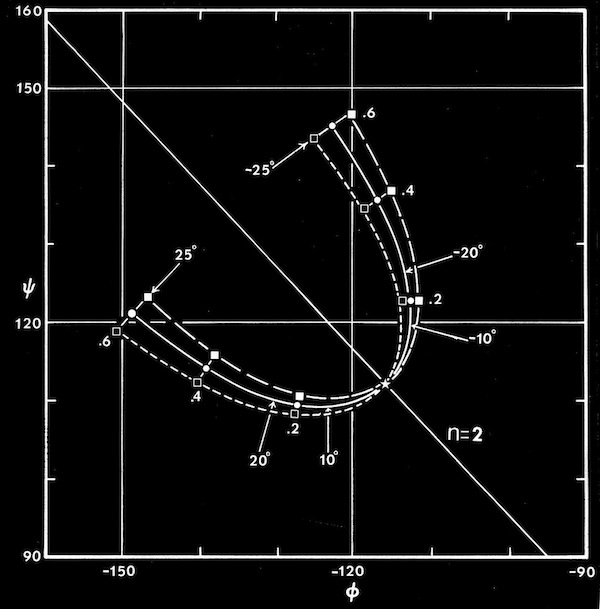
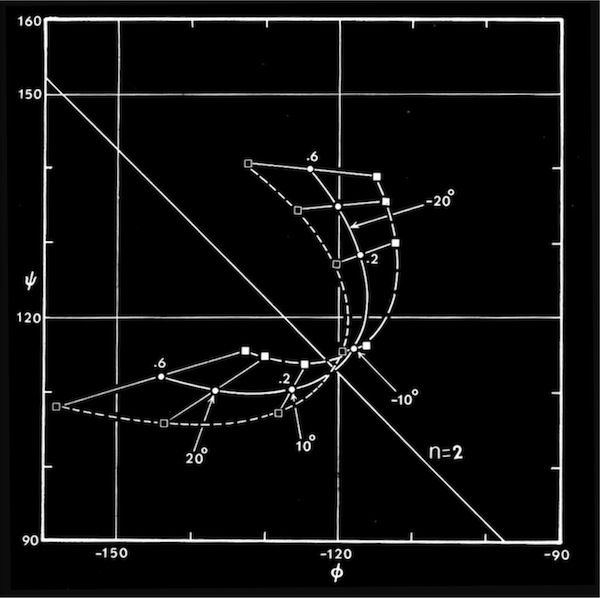
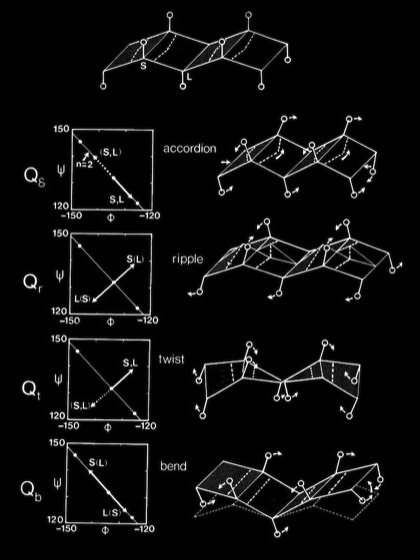
Computational modeling studies showed that idealized flat, parallel beta sheets were composed of polypeptide chains with a defined repeat period of ~ 6.4 Angstroms for a dipeptide with a unique phi,psi conformation lying on the helical n=2 line of the Ramachandran plot. Twisted parallel sheets could be formed of either “crossing” chains (where the phi, psi values for all residues were identical), or from “coiled” chains characterized by alternating pairs of phi,psi conformational angles.
As manifest in the smooth twisting of the mechanical models of parallel beta sheets, the computations showed a uniquely defined trajectory on the Ramachandran plot for both the crossing and coiled H-bonded sheets. The difference between twisted sheets containing crossing vs. coiled polypeptide chains depended on the extent of hydrogen bonding across the sheet. Interior sheet strands, continuously H-bonded to strands on both sides, were ideally constrained to have identical phi,psi values for all residues, while strands at sheet edges (or in double strand sheets) could achieve more highly twisted coiled conformations.
Parallel Beta Sheet Geometry
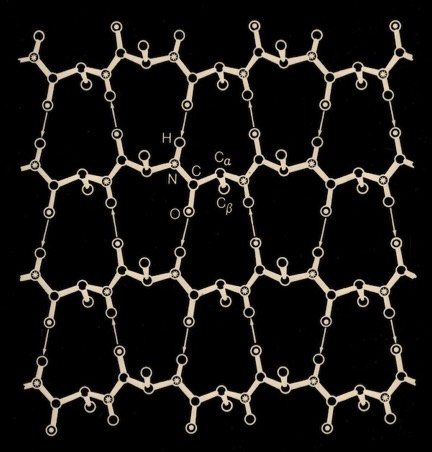
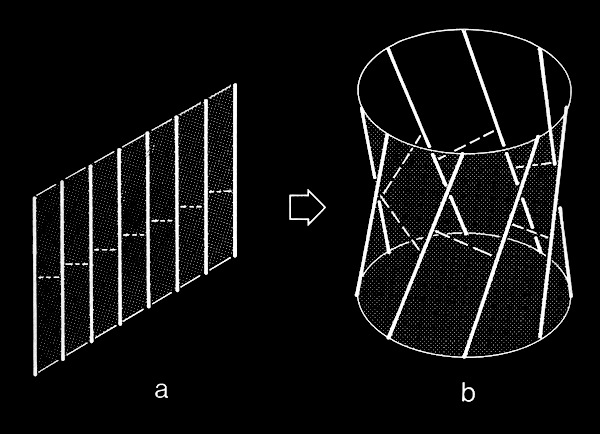
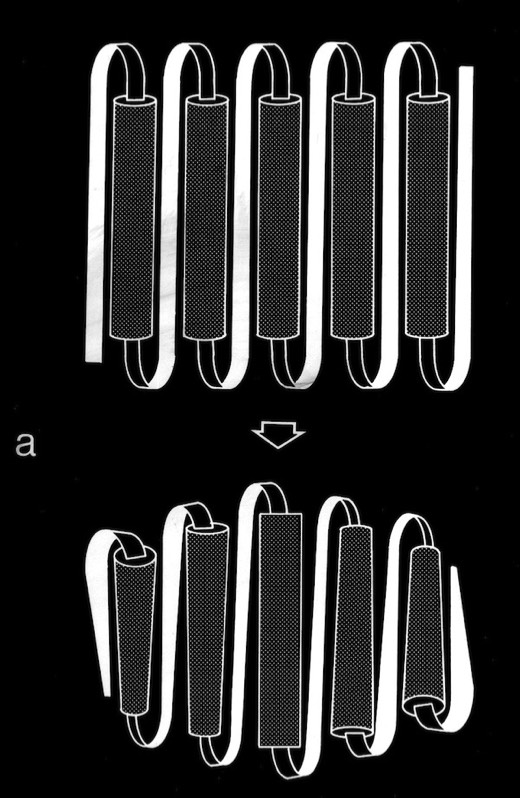
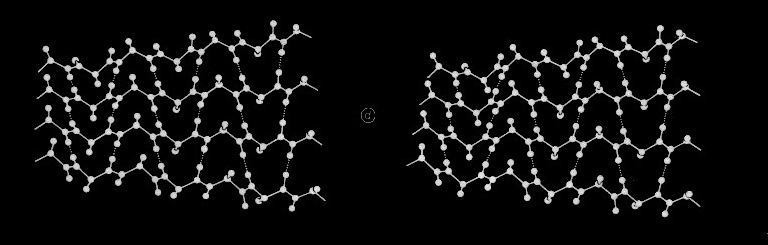
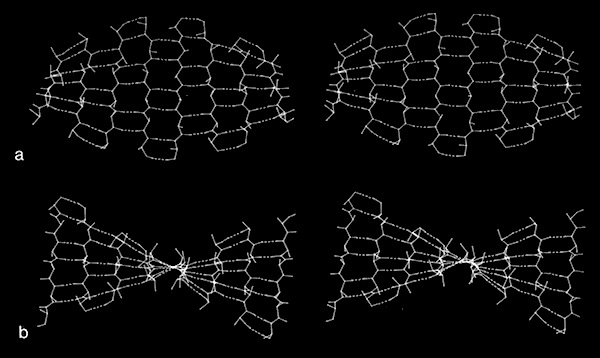
In the course of our work on beta sheets, we developed detailed models for parallel sheets that generally corresponded to structures seen in then known protein structures. A key observation was that the chains in parallel sheets appeared to twist to their greatest extent consistent with the preservation of good interchain H-bonds. However, quite different structures were produced as a consequence of the nature and extent of the pattern of inter-strand H-bonds. For example, sheets with a rectangular overall plan formed twisted minimal surfaces in the form of hyperbolic paraboloids, while sheets with staggered chain arrangements could form parallel beta barrels corresponding to another minimal surface, an hyperboloid of revolution. The obvious structural efficiency and rigidity of these structures is manifest in their application to macroscale structures such as those designed by Frei Otto as well as familiar structures like power plant cooling towers. In the case of the twisted beta sheet structures seen in proteins, the final configurations appear to maximize the extent of chain twist, subject to the geometrical constraints imposed by the organization of the interchain hydrogen bonds, and to approximate minimal anticlastic surfaces.
Parallel beta-barrels, as first observed in Triose Phosphate Isomerase, were shown to be a consequence of polypeptide chain stagger and interchain twist, forming a hyperbol0id of revolution, another anticlastically curved, minimal surface.
The tendency for extended polypeptide chains to assume local left-handed twist also appears to manifest itself in the very frequent occurrence of "right-handed" supercoil loops between protein secondary structures, including the connections in beta-alpha-beta structures noted by Jane Richardson. Coincidently, the twisted beta-alpha-beta structures simultaneously allow both the helices to pack on the sheet and the helices to pack with each other.
The classic beta-alpha-beta barrel in Triose Phosphate Isomerase (1YPI) occurs in many related enzymes from bacteria to humans.
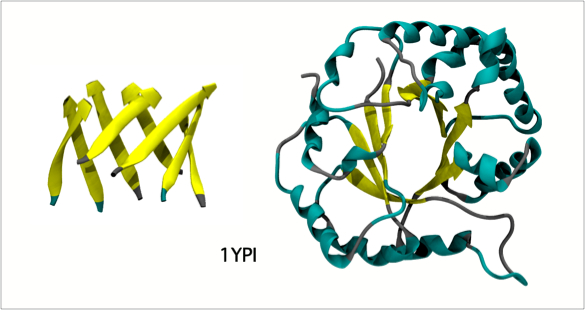
Triose Phosphate Isomerase
Peptide Bond Planarity and Beta Sheet Twist Sense
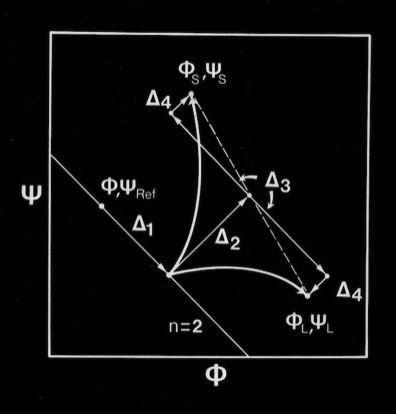
Right-handed supercoiling is a common feature of many recurrent structural features in proteins, and it can be shown that some structures such as double-strand, antiparallel right-twisted coils can achieve lower energy conformations that left-twisted coils. Nevertheless, we were generally able to generate perfectly reasonable looking left-twisted beta sheets, as indicated by the phi,psi trajectories running left from the n=2 line indicated on the accompanying beta sheet Ramachandran plots.
Why then did the polypeptide chains so predominantly twist left? One possibility clearly related to the native chirality of amino acids. In fact, the small molecule X-ray and neutron crystallography available at the time (1977) indicated a preference for chiral distortion at the nitrogen atom of the backbone amide group of L-amino acid peptides. It seemed feasible that polypeptides in solution could have some tetrahedral character (producing a universal tendency in extended polypeptides to form right-handed coils), but then become more planar when forming the extended H-bonded interactions characteristic of alpha-helices and beta sheets. In fact, it turned out that the predominant observed distortion (a positive omega, negative theta nitrogen distortion) turned out to be consistent with optimally H-bonded sheets with a right-handed twist. More recent examinations of peptide bond planarity in highly refined protein structures do not obviously show this trend in omega peptide distortions. Nevertheless, such possibilities may be interesting to reexamine in the context of newer refinement methods and/or specific secondary structures, given the vastly expanded database of ultrahigh resolution X-ray and neutron structures now available.
Why then did the polypeptide chains so predominantly twist left? One possibility clearly related to the native chirality of amino acids. In fact, the small molecule X-ray and neutron crystallography available at the time (1977) indicated a preference for chiral distortion at the nitrogen atom of the backbone amide group of L-amino acid peptides. It seemed feasible that polypeptides in solution could have some tetrahedral character (producing a universal tendency in extended polypeptides to form right-handed coils), but then become more planar when forming the extended H-bonded interactions characteristic of alpha-helices and beta sheets. In fact, it turned out that the predominant observed distortion (a positive omega, negative theta nitrogen distortion) turned out to be consistent with optimally H-bonded sheets with a right-handed twist. More recent examinations of peptide bond planarity in highly refined protein structures do not obviously show this trend in omega peptide distortions. Nevertheless, such possibilities may be interesting to reexamine in the context of newer refinement methods and/or specific secondary structures, given the vastly expanded database of ultrahigh resolution X-ray and neutron structures now available.
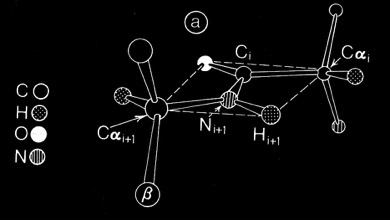
Planar Peptide Bond
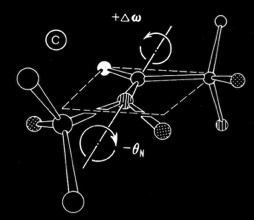
Non-Planar Peptide Bond (+Omega, -Theta)
AntiParallel Beta-Sheets
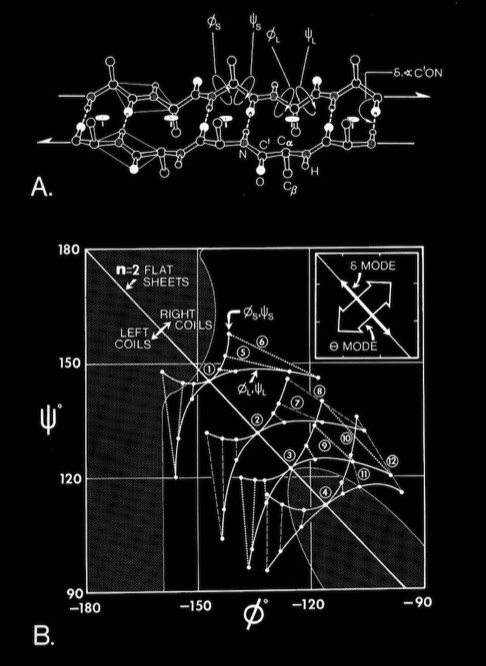
Antiparallel-Sheets: In contrast to parallel sheets where the hydrogen-bonded “rings” formed between adjacent strands are all equivalent, antiparallel sheets form a pattern of alternating "small" and "large" hydrogen bonded rings and incorporate additional dyad symmetry elements normal to the plane of the sheet. In our computational modeling studies, we assumed that H-bonds of equivalent length where the N-H vector lay in the plane of the peptide carbonyl oxygen sp2 orbital were essentially equivalent energetically. This possibility admits a range of flat antiparallel sheets, all with conformations lying along the n=2 line of the Ramachandran plot, but with slightly different interchain H-bond geometry and dipeptide repeat periods.
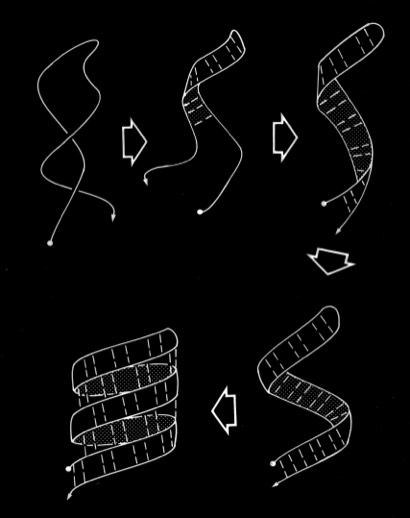
Double-strand antiparallel sheets can twist with essentially perfect preservation of the interchain H-bonds if the chains form coils with dipeptide repeating units. The progressive coiling in double-stranded antiparallel sheets is highly cooperative, allowing them to perform as a flexible structural element in many protein conformational changes.
Antiparallel Double Strand Coil Description
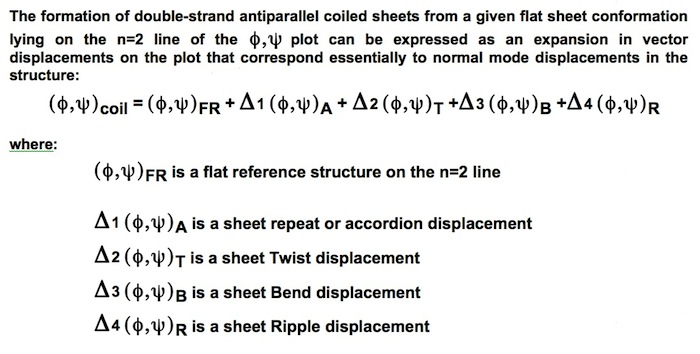
The degrees of flexibility present in antiparallel beta sheets allows them to form a variety of geometrical conformations included highly twisted Hyperbolic Paraboloid conformations. In this model, the HP sheet is composed of identical straight helical chains with phi = -122 degrees, psi = 135 degrees, and a slightly non-linear interchain H-bond angle delta of 170 degrees.
Antiparallel HP Sheet
The ability of antiparallel sheets to form highly twisted sheets or other unusual surfaces is a result of the inherent flexibility of the antiparallel coiled conformations.
Antiparallel Domed Sheet Coiled Chains
Antiparallel coiling flexibility also accounts for the alternate geometry observed for the antiparallel barrels illustrated at the top of this page, whose structures were not known at the time of our initial analysis.
Antiparallel Beta-Sheet Folding
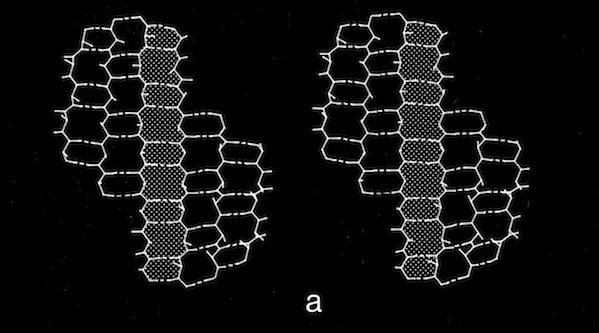
The coiling characteristics of double-strand antiparallel sheets connected by a hairpin bends are of particular interest as possible intermediates in protein folding. One feature to emerge from examination of structures composed of antiparallel beta sheets, noted particularly by Jane Richardson in her descriptions of antiparallel beta barrel topology, was the recurrence of "Greek Key" motifs in the H-bonding plan of antiparallel beta sheets and barrels. Although there is quite a bit of variation in the H-bond plan of Greek-Key sheets, Richardson suggested the plausibility of folding such structures via antiparallel beta ribbon intermediates comprising two antiparallel beta sheet strands linked by a hairpin bend. Our analysis of the conformational flexibility of double strand sheets shows them able to flexibly coil while retaining perfectly intact H-bonds, thus demonstrating the physical feasibility of these models in protein folding.
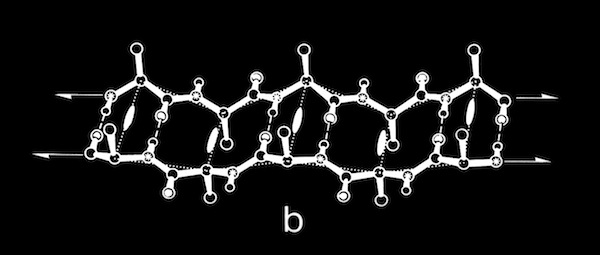
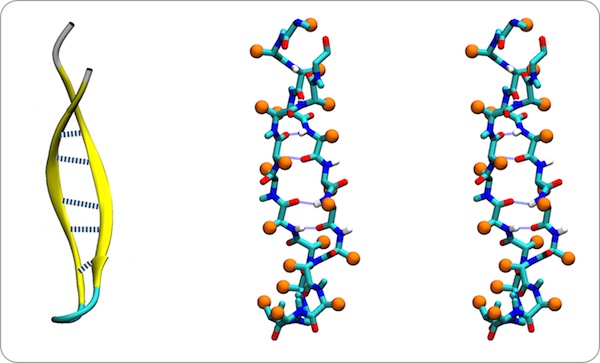
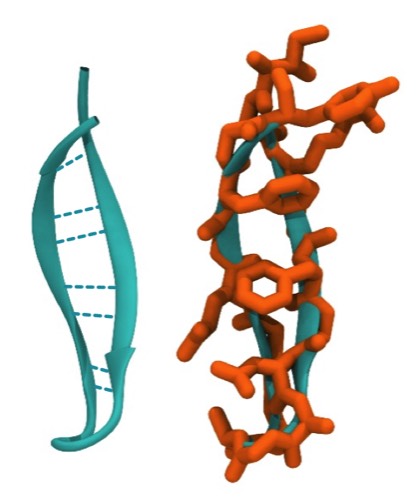
In addition to the intrinsic flexibility of the antiparallel coil, we recognized an additional property of the coiled structure that we felt could assist in understanding how these structures might fold. This stems from the observation that the sides of double strand antiparallel coils are different, and that right- twisted coils fold with the residues situated in small H-bonded rings oriented toward the coil axis. This is a feature, for example in the PTI beta sheet.
This sidedness feature turns up in some multi-strand antiparallel beta sheet structures, but not in others. This then begs the question whether the whole idea is wrong, is somehow determined by specifics of the closing beta turn residues, or otherwise implies that the structure rearranges after getting close to a finally folded form. It is known for example, that some beta sheet proteins can be cyclically permuted and still fold into functional structures.
Antiparallel Beta-Bulge
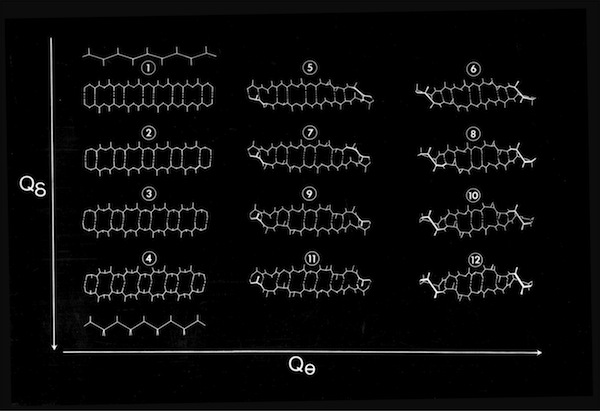
An additional manifestation of the flexibility in antiparallel sheets turns up in the modeling of triple strand structures. In some contexts, where sheets were packed on other sheets, sheets conformed to an expected pattern where the central, strand approximated a straight twisted conformation (all phi,psi conformational angles equivalent) and the outer strands approximated more highly twisted coiled conformations. However, double strand sheets can become highly coiled to an extent that allows amino acid side chains to pack in interdigitating fashion, forming a structural minidomain with a compact structural core. It is not possible for a third polypeptide chain continuously H-bond to a highly twisted coiled chain, so a hairpin-like bend or “beta-bulge” is inserted in the exterior chain to accommodate the longer contour followed by the exterior chain.
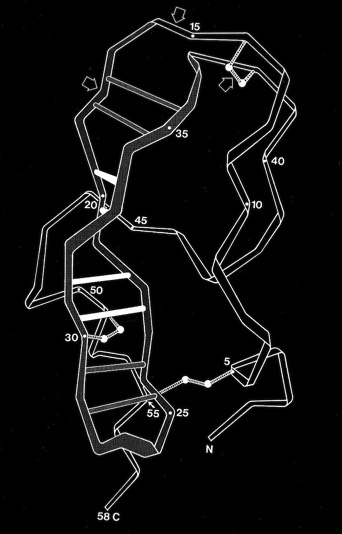
PTI and Coiled-Coil H-Bond Stability
Double-strand antiparallel sheets connected by a hairpin bends can form highly twisted coils where residues projecting from the inside surface can pack to form a structural minidomain with a compact structural core. An example is the double stranded beta-coil in pancreatic trypsin inhibitor.
NMR measurements of the amide proton exchange rates in PTI showed that the relatively solvent exposed amide protons involved in the beta sheet H-bonds were unusually resistant to exchange, an observation that seemed contrary to the more usual observation that H-exchange rates tended to be highly correlated with solvent accessibility. I pointed out that the H-exchange stability might be explained in part by the ability of the antiparallel beta coil to undergo correlated conformational transitions from the ground state without necessarily breaking any sheet H-bonds.
NMR measurements of the amide proton exchange rates in PTI showed that the relatively solvent exposed amide protons involved in the beta sheet H-bonds were unusually resistant to exchange, an observation that seemed contrary to the more usual observation that H-exchange rates tended to be highly correlated with solvent accessibility. I pointed out that the H-exchange stability might be explained in part by the ability of the antiparallel beta coil to undergo correlated conformational transitions from the ground state without necessarily breaking any sheet H-bonds.