Publications
I was first introduced to the use of thermodynamic methods for protein characterization while working for Fred Richards, who worked with Julian Sturtevant at Yale in the 1960's. Later, at Genex, when I became involved in engineering proteins with enhanced stability for industrial applications, I worked with Mike Pantoliano and Jim Matthew with early DSC instruments to characterize engineered forms of subtilisin. At DuPont we applied both ITC and DSC approaches to characterizing ligand binding of native and structure-based ligands to streptavidin.
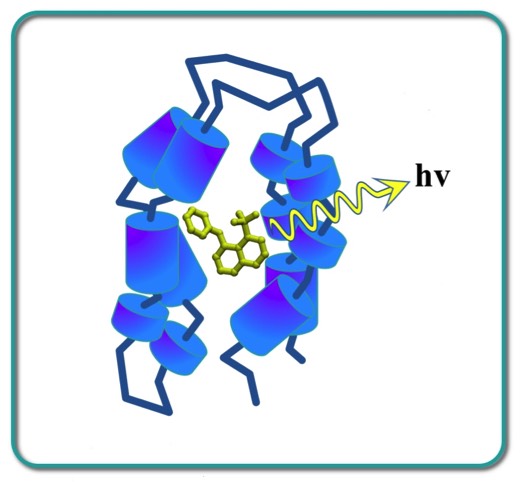
At 3-Dimensional Pharmaceuticals (3DP), Mike Pantoliano developed a basic technique using ANS, which fluoresces strongly when bound to thermally denatured proteins, to develop a high-throughput, miniaturized format to measure the melting point of proteins. The technology and custom instrumentation was extensively developed at 3DP as the ThermoFluor platform for a wide variety of protein characterization and label-free HTS applications.
In recent years, the advent of widely available qPCR instrumentation and alternative fluorescent dyes has lead to the wide adoption of the thermofluor method, variously referred to as "differential scanning fluorimetry" , "fluorescence thermal shift assay", or "temperature dependent fluorescence" (although this latter term refers to many different phenomena).
At 3-Dimensional Pharmaceuticals (3DP), Mike Pantoliano developed a basic technique using ANS, which fluoresces strongly when bound to thermally denatured proteins, to develop a high-throughput, miniaturized format to measure the melting point of proteins. The technology and custom instrumentation was extensively developed at 3DP as the ThermoFluor platform for a wide variety of protein characterization and label-free HTS applications.
In recent years, the advent of widely available qPCR instrumentation and alternative fluorescent dyes has lead to the wide adoption of the thermofluor method, variously referred to as "differential scanning fluorimetry" , "fluorescence thermal shift assay", or "temperature dependent fluorescence" (although this latter term refers to many different phenomena).
78. Crystallographic and Thermodynamic Comparison of Structurally Diverse Molecules Binding to Streptavidin.
P.C. Weber, M.W. Pantoliano, F.R. Salemme Acta Cryst 1995 D51, 590-596
P.C. Weber, M.W. Pantoliano, F.R. Salemme Acta Cryst 1995 D51, 590-596
89. High Density Miniaturized Thermal Shift Assay as a General Strategy for Drug Discovery.
M.P. Pantoliano, E. Petrella, J. Kwasnoski, V. Lobannov, J. Myslik, E. Graf, T. Carver, E. Asel, B. Springer, F.R. Salemme Journal of Biomolecular Screening. 2001; 6(6):492-440
M.P. Pantoliano, E. Petrella, J. Kwasnoski, V. Lobannov, J. Myslik, E. Graf, T. Carver, E. Asel, B. Springer, F.R. Salemme Journal of Biomolecular Screening. 2001; 6(6):492-440
Original thermofluor assay description (More here).
91. Applications of Calorimetric Methods to Drug Discovery and the Study of Protein Interactions.
P.C. Weber & F.R. Salemme, Curr. Opinion in Structural Biol. 2003; 13:115-121
P.C. Weber & F.R. Salemme, Curr. Opinion in Structural Biol. 2003; 13:115-121
92. Direct Binding Assays for Pharma Screening.
M.J. Todd & F.R. Salemme, Genetic Engineering News. 2003;3:28-29
M.J. Todd & F.R. Salemme, Genetic Engineering News. 2003;3:28-29
94. Decrypting the Biochemical Function of an Essential Gene from Streptococcus pneumoniae using Thermofluor Technology.
Carver TE, Bordeau B, Cummings MD, Petrella EC, Pucci MJ, Zawadzke LE, Dougherty BA, Tredup JA, Bryson JW, Yanchunas J Jr, Doyle ML, Witmer MR, Nelen MI, Desjarlais RL, Jaeger EP, Devine H, Asel ED, Springer BA, Bone R, Salemme FR, Todd M. J Biol Chem. 2005; 280:11704-11712
Carver TE, Bordeau B, Cummings MD, Petrella EC, Pucci MJ, Zawadzke LE, Dougherty BA, Tredup JA, Bryson JW, Yanchunas J Jr, Doyle ML, Witmer MR, Nelen MI, Desjarlais RL, Jaeger EP, Devine H, Asel ED, Springer BA, Bone R, Salemme FR, Todd M. J Biol Chem. 2005; 280:11704-11712
3DP ran HTS screening campaigns using thermofluor on many targets identified through gene knockout studies that were otherwise poorly characterized with respect to chemical function. We quickly developed a strategy of prescreening each protein with a "functional probe library" of about 3000 biologically relevant compounds including metal ions, cofactors, fragments and enzyme inhibitor. The binding effects often facilitated a higher level of sequence comparison than possible using a BLAST sequence search, that in turn allowed functional identification of the target.
95. Thermodynamic Stability Of Carbonic Anhydrase: Measurements Of Binding Affinity And Stoichiometry Using Thermofluor.
Matulis D, Kranz JK, Salemme FR, & Todd MJ Biochemistry 2005; 44(13): 5258-66.
Matulis D, Kranz JK, Salemme FR, & Todd MJ Biochemistry 2005; 44(13): 5258-66.