4-Alpha Helical Bundle Proteins
Cyotochrome C-prime
The determination of cytochrome c-prime (PDB Code 2CCY), a prototypical 4-alpha helical bundle protein, motivated studies that first described this recurrent protein structural motif and explained how it was incorporated into several different structural and functional contexts in which it had been observed. At the time of this work (1980) there were less than 100 3D structures deposited in the Protein Data Bank, at that time housed at the Brookhaven National Laboratory
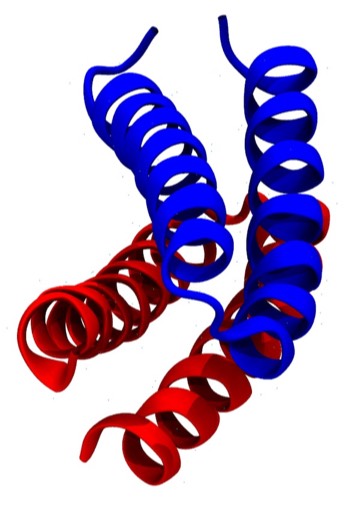
Crick first proposed the general features of “knobs-into-holes” side chain packing interactions between coiled-coil alpha helices for fibrous alpha helical proteins in 1953. However, alpha helical proteins like myoglobin and many others incorporating relatively short alpha helices seldom manifest helices that are obviously coiled. One exception is the ROP protein (PDB Code 1GTO), which is an unusually symmetric dimer whose helices form an antiparallel coiled-coil 4-helical bundle. The ROP protein differs from most single-chain 4-alpha helical bundle proteins, which generally incorporate relatively straight helices and form bundles with an hourglass or wedge-like overall shape
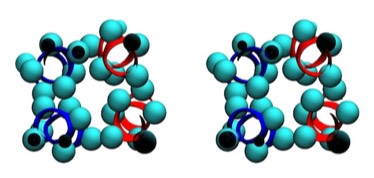
ROP Cross Section
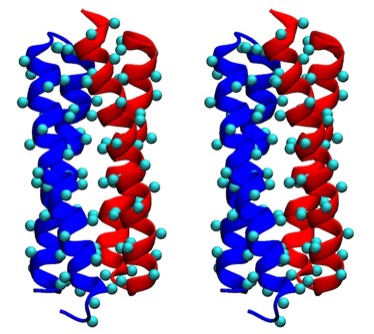
ROP Cross Stereo
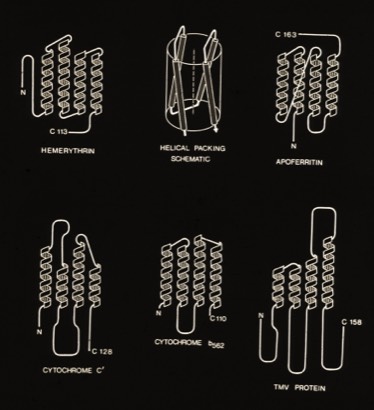
4-Alpha Helix Bundle Proteins
(Circa 1980)
The protein structures examined in the original description of the 4-alpha helical bundle motif in 1980 manifest little apparent sequence homology and were generally constructed of relatively short alpha helices. Nevertheless, it was clear that the most compact arrangement of 4 alpha helices with similar lengths and short interconnections was an antiparallel bundle. The most symmetric arrangements had an “hourglass” shape with points of closest inter-helical approach at the centers of each helix. However, most observed structures had a point of closest interhelical approach at one end of the bundle, making them wedge-shaped.
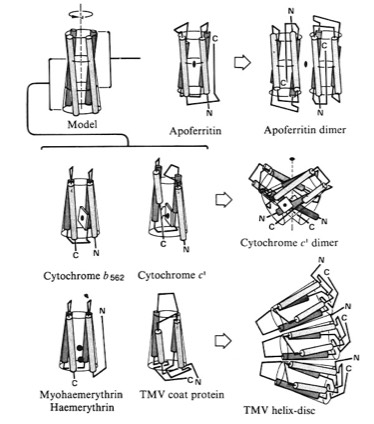
4-Alpha Helical Bundle Superstructures
The more symmetric 4-alpha helical bundle arrangements, with an “hourglass” shape, were suited for packing into correspondingly symmetric superstructures like ferritin. Structures with the point of interhelix closest approach at one end of the bundle that were wedge-shaped could pack to form discs or helical arrays such as found in TMV. Alternately, the divergent end of a wedge-shaped bundle could form an internal ligand binding site, as it did for cytochrome b562 and cytochrome c-prime.
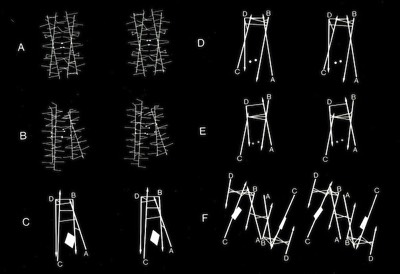
4-Alpha Helix Bundle Interhelical Contacts
Assessments of alpha helix packing in proteins were developed as an extension of the original “knobs-into-holes” model for alpha-helical coiled coils originally advanced by Crick in 1953 in order to develop models for geometrically preferred interhelical interactions. The approach basically involved finding alternative superpositions of two alpha helix nets that could reflect possible patterns of periodic interdigitization of the side chains of the two alpha helices. The approach only provides an approximation for straight alpha helices which rapidly diverge from their points of closest approach in 4-helical bundles.
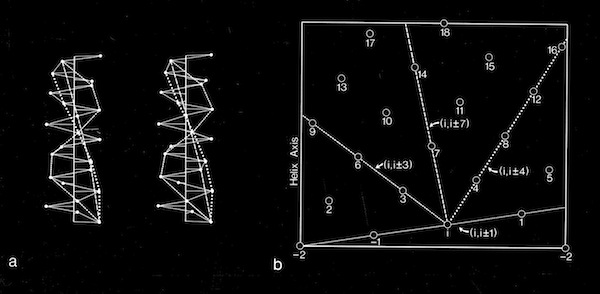
Alpha Helix Net (after Crick 1953)
For interactions between straight helices, that can be alternative patterns of local side chain interdigitation at interhelical angles of ~20 degrees as seen in 4-alpha helical bundles.
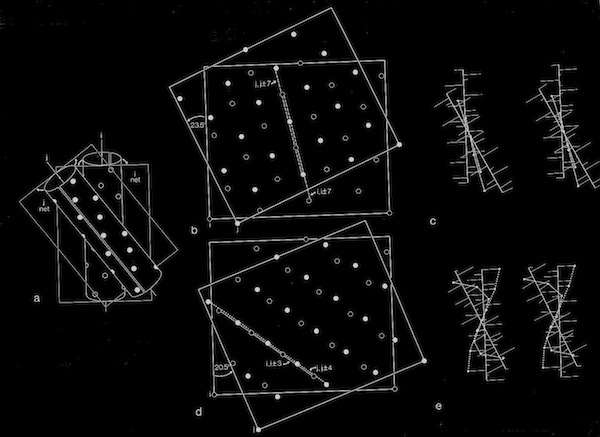
Alpha Helix Interactions at ~20 Degrees
Models of helix interactions in 4-alpha helical bundles at ~18-20 degrees admit a variety of possible bundle pseudosymmetries and corresponding interhelical packing interactions, many of which incorporate localized favorable packing interactions. The frequency of recurrence of the 4-alpha helix bundle motif evidently stems from the great variety of alternatively packed arrangements that can nevertheless form locally favorable packing interactions within a common overall geometrical framework.
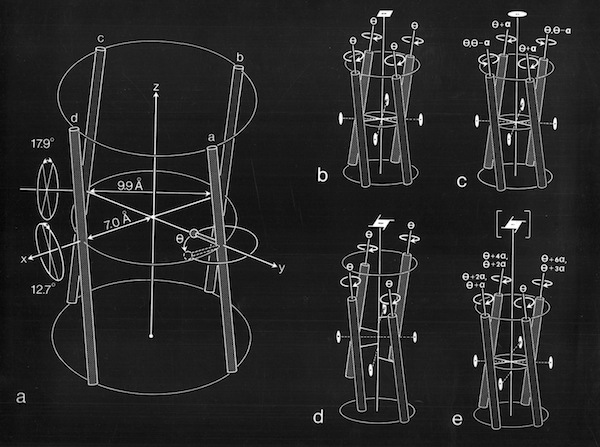
4 Alpha Helix Bundle Geometry & Packing Symmetry
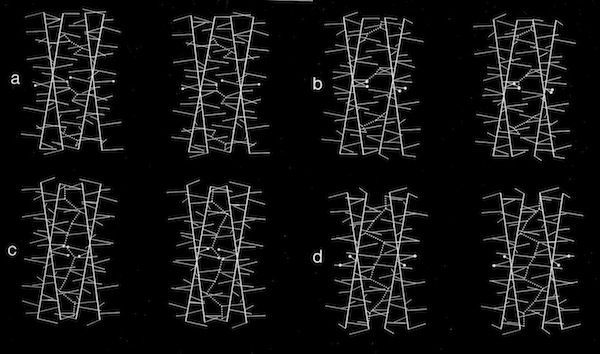
Stereo Views of Model 4 Helix Bundle Packing at 18 Degrees
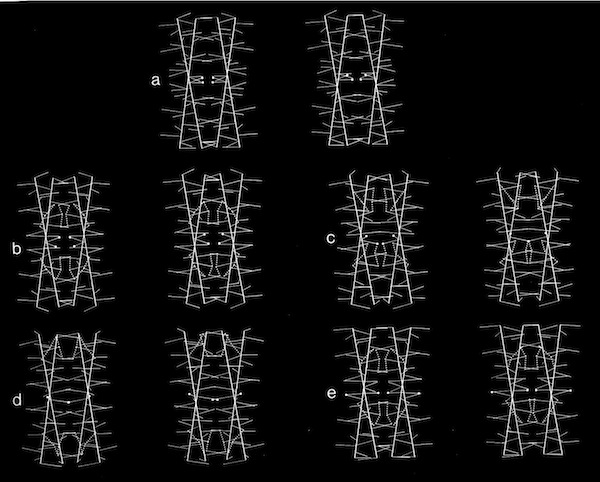
Dyad Related 4 Helix Bundle Packing
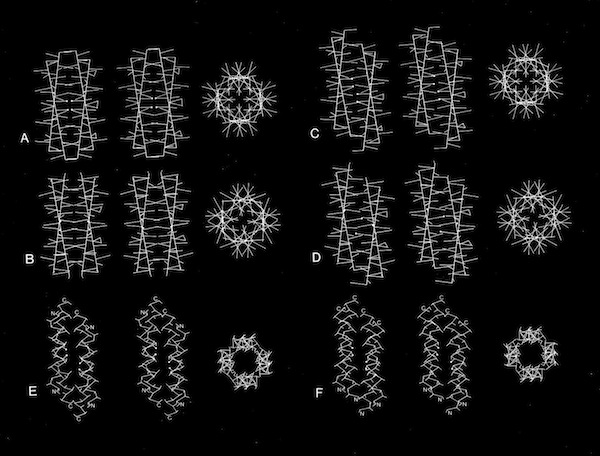
4(1) Screw 4-Helix bundle Packing
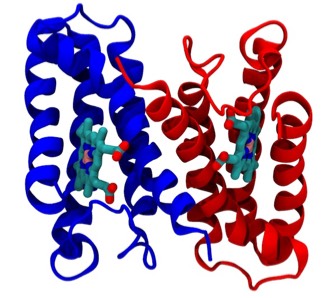
The cytochrome b562 from E. coli was observed to be a 4-alpha helical bundle protein with architecture similar to monomer of the cytochrome c-prime derived from the photosynthetic bacterium Rhodopseudomas molischianum. This was a bit surprising since the protoporphyrin IX heme group in cytochrome c-prime is covalently connected to the polypeptide backbone through thioether linkages to protein cysteine groups (a defining property of c-type cytochromes), whereas the heme was non-covalently bound in cytochrome b 562. Nevertheless, a least square superposition of the heme groups in the two structures showed a surprising correspondence between the both the alpha helical frameworks of the two structures as well as the situation of several aromatic amino acid side chains surrounding the heme groups. Is this a manifestation of convergent or divergent evolution?
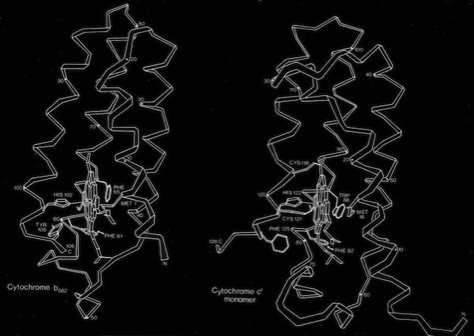
Cytochrome b562 and Cytochrome c-Prime
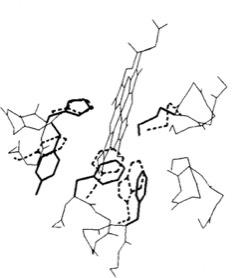
Heme Superposition
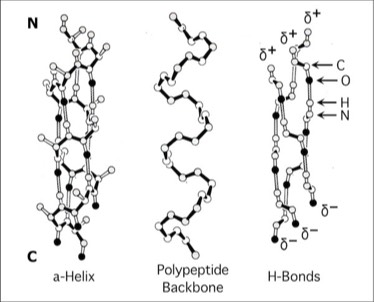
Alpha Helix Macro Dipoles
The contiguous H-bond networks that stabilize the alpha helical conformation create a helical macrodipole.
In a collaborative study with Bob Sheridan and Ron Levy, interaction of the helix macrodipoles were determined to be an energetically favorable contributor to the antiparallel helix arrangements found both in 4-alpha helical bundle proteins and their aggregated superstructures.

H-Bond Networks in Cytochrome c-Prime Dimer
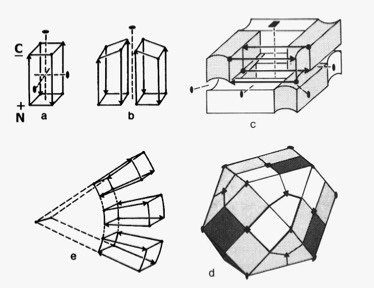
Dipolar Stabilization in 4-Alpha Helical Superstructures