Publications
I became interested in science and mechanical systems at a young age and was fortunate to have been trained as a machinist by a friend of the family who was an extraordinarily gifted tool and die maker. I was accepted at MIT and Yale without finishing high school. On attending Yale, I was introduced to Fred Richards and Hal Wyckoff, who were among the first US scientists to develop capabilities for protein crystallography. Both Fred and Hal liked to invent and continuously modify instrumentation. Although Yale had outstanding professional machinists supporting researchers, I was able to quickly evolve prototype designs to fit the always changing experimental requirements (e.g. "We need a hole drilled here."). Subsequently I applied my mechanical skills to the construction and development of numerous pieces of hardware related to biophysics research and protein crystallography and retain a strong interest in the development of new instrumentation for protein crystallography and other applications.
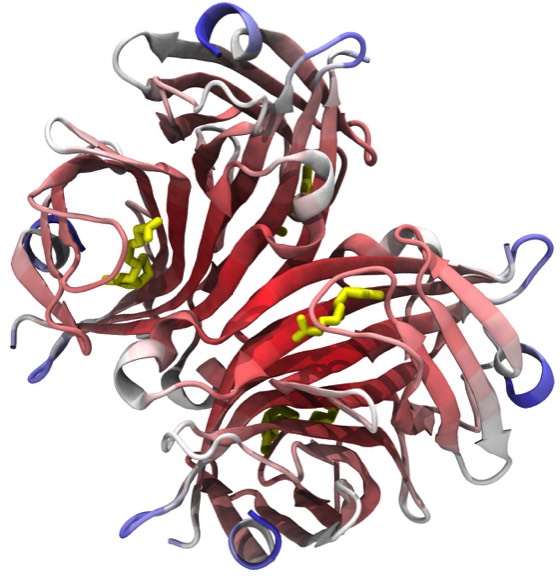
0. The Crystal Structure of Cytochrome C2 from R. rubrum and Related Topics in Protein Crystallography.
F. R Salemme 1972 (UCSD PhD Thesis)
F. R Salemme 1972 (UCSD PhD Thesis)
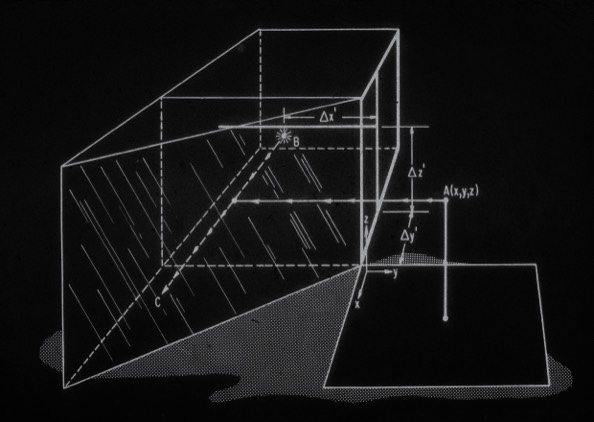
2. A Device for the Rapid Measurement of Molecular Model Coordinates for X-ray Crystallography.
F.R. Salemme & D.G. Fehr J. Mol. Biol. 1972; 70: 697-700
F.R. Salemme & D.G. Fehr J. Mol. Biol. 1972; 70: 697-700
Prior to the advent of computer graphics for construction of protein models from electron density maps, physical models were constructed using Kendrew wire models. The models were constructed based on optical superposition (using a Richards Optical Comparator, which was inspired by the Leica M-series optical rangefinder) with a sectioned electron density map. Coordinates were read off the assembled model with a grid and plum bob, a highly inaccurate and tedious process. Dave Fehr and I designed and built a mechanical device that used the optical comparator to superimpose a small light on the reflected image of the assembled molecular model and simultaneously provide a digital coordinate readout. The device enabled pioneering early efforts in protein refinement by Richard Alden and Steven Freer in the UCSD Kraut lab.
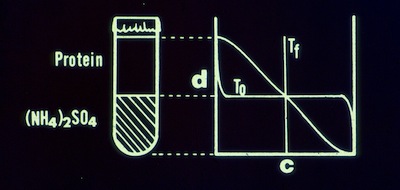
3. A Free Interface Diffusion Technique for the Crystallization of Proteins for X-ray Crystallography.
F.R. Salemme, Arch. Biochem. Biophys. 1972; 151: 533-539.
F.R. Salemme, Arch. Biochem. Biophys. 1972; 151: 533-539.
I introduced the use of free interface diffusion for protein crystallization. The method creates a highly supersaturating condition at the initial interface that induces nucleation, then gradually relaxes the local protein and precipitant concentrations to allow a few surviving nucleii to grow to large crystals. The method is not as popular as hanging or sitting drop methods that are more readily suited to automation. Nevertheless FID creates transient conditions than are different from other methods, and it still remains a useful approach to protein crystallization.
34. Modeling Correlated Motions in Proteins and Implications for Protein Refinement.
F.R. Salemme and C.E. KenKnight Transactions of the American Crystallographic Association, 1984; 20: 97-108
F.R. Salemme and C.E. KenKnight Transactions of the American Crystallographic Association, 1984; 20: 97-108
36. Protein Crystallization by Free Interface Diffusion.
F.R. Salemme Methods in Enzymology (S. Timasheff and H. Wyckoff, Eds.) 1985; 114: 140-141
F.R. Salemme Methods in Enzymology (S. Timasheff and H. Wyckoff, Eds.) 1985; 114: 140-141
38. Anomalous Temperature Factor Behavior and Crystal Lattice Mobility in Cytochrome c’.
B.C. Finzel and F.R. Salemme Biophys. J. 1985; 49: 73-76
B.C. Finzel and F.R. Salemme Biophys. J. 1985; 49: 73-76
41. Lattice Mobility and Anomalous Temperature Factor Behavior in Cytochrome c’.
B.C.Finzel and F.R.Salemme, Nature 1985; 315: 686-688.
B.C.Finzel and F.R.Salemme, Nature 1985; 315: 686-688.
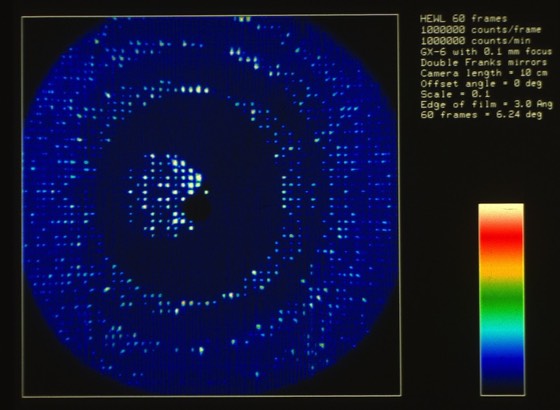
47. Use of an Imaging Proportional Counter in Macromolecular Crystallography.
A.J. Howard, G.L. Gilliland, B.C. Finzel, T.L. Poulos, D.H. Ohlendorf, and F.R. Salemme, J. Appl. Cryst. 1987; 20: 383-387
A.J. Howard, G.L. Gilliland, B.C. Finzel, T.L. Poulos, D.H. Ohlendorf, and F.R. Salemme, J. Appl. Cryst. 1987; 20: 383-387
48. Molecular Factors Stabilizing Protein Crystals.
F.R.Salemme, Lars Genieser, B.C. Finzel, R.M. Hilmer, J.J. Wendoloski, J. Crystal Growth 1988; 90: 273-282.
F.R.Salemme, Lars Genieser, B.C. Finzel, R.M. Hilmer, J.J. Wendoloski, J. Crystal Growth 1988; 90: 273-282.
51. Protein Crystal Growth in Microgravity.
L.J. DeLucas, C.D. Smith, H.W. Smith, V.K. Senadhi, S.E. Senadhi,S.E. Ealick, D.C. Carter, R.S., Snyder, P.C. Weber, F.R. Salemme, D.H. Ohlendorf, H.M. Einspahr, L.L. Clancy, M.A. Navia, B.M McKeever, T.L. Nagabhushan, G. Nelson, A. McPherson, S. Koselak, G. Taylor, D. Stammers, K. Powell, G. Darby, C.E. Bugg Science 1989; 246: 651-654.
L.J. DeLucas, C.D. Smith, H.W. Smith, V.K. Senadhi, S.E. Senadhi,S.E. Ealick, D.C. Carter, R.S., Snyder, P.C. Weber, F.R. Salemme, D.H. Ohlendorf, H.M. Einspahr, L.L. Clancy, M.A. Navia, B.M McKeever, T.L. Nagabhushan, G. Nelson, A. McPherson, S. Koselak, G. Taylor, D. Stammers, K. Powell, G. Darby, C.E. Bugg Science 1989; 246: 651-654.
54. Protein Crystal Growth in Microgravity.
L.J. DeLucas, C.D. Smith, H.W. Smith, S. Vijay-Kumar, S.E. Senadhi, S.E. Ealick, C.E. Bugg, D.C. Carter, R.S., Snyder, P.C. Weber, F.R. Salemme, D.H. Ohlendorf, H.M. Einspahr, L.L. Clancy, M.A. Navia, B.M McKeever, T.L. Nagabhushan, G. Nelson, A. McPherson, S. Koszelak, G. Taylor, D. Stammers, K. Powell, G. Darby Proceedings VIIth European Symposium on Materials and Fluid Sciences in Microgravity 1990; ESA SP-295: 31-34
L.J. DeLucas, C.D. Smith, H.W. Smith, S. Vijay-Kumar, S.E. Senadhi, S.E. Ealick, C.E. Bugg, D.C. Carter, R.S., Snyder, P.C. Weber, F.R. Salemme, D.H. Ohlendorf, H.M. Einspahr, L.L. Clancy, M.A. Navia, B.M McKeever, T.L. Nagabhushan, G. Nelson, A. McPherson, S. Koszelak, G. Taylor, D. Stammers, K. Powell, G. Darby Proceedings VIIth European Symposium on Materials and Fluid Sciences in Microgravity 1990; ESA SP-295: 31-34
67. Protein Crystal Growth Results for Shuttle Flights STS-26 and STS-29.
L.J. DeLucas, C.D. Smith, H.W. Smith, V.K. Senadhi, S.E. Ealick, D.C. Carter, R.S., Snyder, P.C. Weber, F.R. Salemme, D.H. Ohlendorf, H.M. Einspahr, L.L.Clancy,M.A. Navia, B.M McKeever, T.L. Nagabhushan, G. Nelson, A. McPherson, S. Koselak, G. Taylor,D. Stammers, K. Powell, G. Darby, and C.E. Bugg. J. Crystal Growth 1991; 110: 302-311.
L.J. DeLucas, C.D. Smith, H.W. Smith, V.K. Senadhi, S.E. Ealick, D.C. Carter, R.S., Snyder, P.C. Weber, F.R. Salemme, D.H. Ohlendorf, H.M. Einspahr, L.L.Clancy,M.A. Navia, B.M McKeever, T.L. Nagabhushan, G. Nelson, A. McPherson, S. Koselak, G. Taylor,D. Stammers, K. Powell, G. Darby, and C.E. Bugg. J. Crystal Growth 1991; 110: 302-311.